Introduction of SPEED
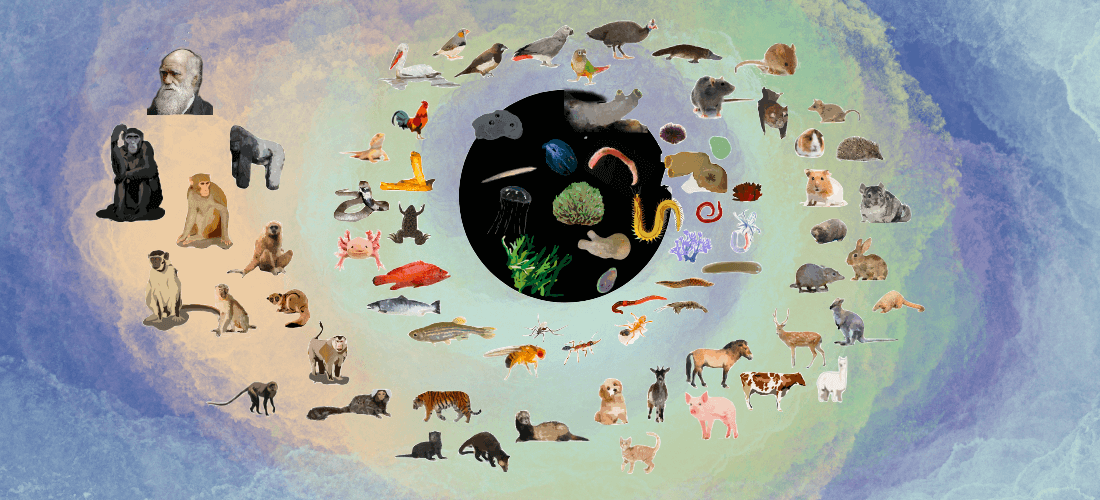
127 species
6,310,504 cells
85 diseases
Statistics of SPEED
Number of species
Number of projects
Number of time points
Number of diseases
Contact us
Phone: +86-512-62873780
Fax: +86-512-62873779
Email: cds@ism.pumc.edu.cn
Address: 100 Chongwen Road, Suzhou Industrial Park, Suzhou, China, 215123
Cite (Please consider citing this study, if SPEED was used):Yangfeng Chen, Xingliang Zhang, Xi Peng, Yicheng Jin, Peiwen Ding, Jiedan Xiao, Changxiao Li, Fei Wang, Ashley Chang, Qizhen Yue, Mingyi Pu, Peixin Chen, Jiayi Shen, Mengrou Li, Tengfei Jia, Haoyu Wang, Li Huang, Guoji Guo, Wensheng Zhang, Hebin Liu, Xiangdong Wang, Dongsheng Chen, SPEED: Single-cell Pan-species atlas in the light of Ecology and Evolution for Development and Diseases, Nucleic Acids Research, 2022;, gkac930, https://doi.org/10.1093/nar/gkac930